Docker EE GPU for Apt Installation Recipe
These are the steps to installing OmniSci as a Docker container on a machine running with NVIDIA Kepler or Pascal series GPU cards.
Preparation
Prepare your host by installing NVIDIA drivers, Docker, and NVIDIA runtime.
Install NVIDIA Drivers
To install NVIDIA drivers, open a terminal window on the host. Run apt update
and apt upgrade to ensure that you are using the latest operating
system software.
sudo apt update sudo apt upgrade
Use apt-get to install required utilities.
sudo apt-get install \
apt-transport-https \
ca-certificates \
curl \
gnupg-agent \
software-properties-common
Reboot your system to activate all of your changes.
sudo reboot
Install CUDA
Verify that the gcc compiler is installed with the following command.
gcc --version
If no version information returns, run the following command.
sudo apt install gcc
To install the CUDA package:
- Go to https://developer.nvidia.com/cuda-downloads.
- Select the target platform by selecting the operating system (Linux), architecture (based on your environment), distribution (Ubuntu), version (based on your environment), and installer type (OmniSci recommends deb (network)).
- Install the CUDA drivers per instructions on the NVIDIA website.
- Reboot your system to ensure that all changes are active:
sudo reboot
Install Docker
Remove any existing Docker installs and the legacy NVIDIA docker runtime.
sudo docker volume ls -q -f driver=nvidia-docker | xargs -r -I{} -n1 docker ps -q -a -f volume={} | xargs -r docker rm -f
sudo apt-get purge nvidia-docker
Remove Docker.
sudo apt-get remove docker docker-engine docker.io containerd runc
Update with apt-get.
sudo apt-get update
Use curl to download the latest Docker version.
sudo curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add -
Add Docker to your Apt repository.
sudo add-apt-repository \ "deb [arch=amd64] https://download.docker.com/linux/ubuntu \ $(lsb_release -cs) \ stable"
Update your repository.
sudo apt-get update
Install Docker, the command line interface, and the container runtime.
sudo apt-get install docker-ce docker-ce-cli containerd.io
Optional: Run the following usermod command so that docker command execution does not require sudo privilege.
Log out and log back in for the changes to take effect.
sudo usermod -aG docker $USER
Checkpoint
Verify your Docker installation.
sudo docker run hello-world
Install NVIDIA Docker Runtime
Use curl to add a gpg key:
curl -s -L https://nvidia.github.io/nvidia-container-runtime/gpgkey | \ sudo apt-key add - distribution=$(. /etc/os-release;echo $ID$VERSION_ID)
Update your sources list:
curl -s -L https://nvidia.github.io/nvidia-container-runtime/$distribution/nvidia-container-runtime.list | \ sudo tee /etc/apt/sources.list.d/nvidia-container-runtime.list
Update apt-get and install nvidia-container-runtime:
sudo apt-get update sudo apt-get install -y nvidia-container-runtime
Edit /etc/docker/daemon.json to add the following, and save the changes:
{
"default-runtime": "nvidia",
"runtimes": {
"nvidia": {
"path": "/usr/bin/nvidia-container-runtime",
"runtimeArgs": []
}
}
}
Restart the Docker daemon:
sudo pkill -SIGHUP dockerd
Checkpoint
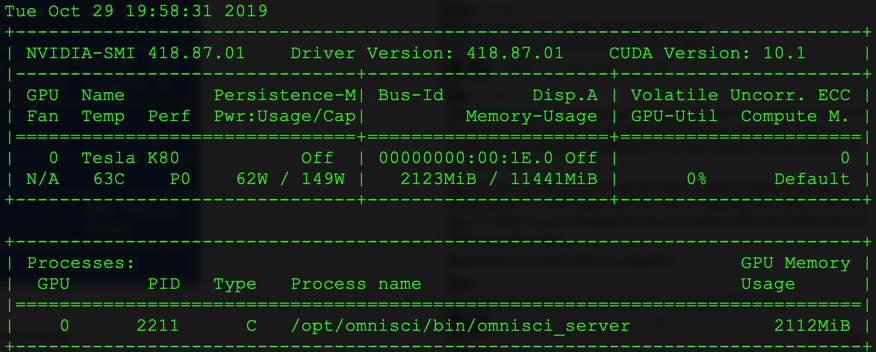
Verify docker and NVIDIA runtime work together.
sudo docker run --runtime=nvidia --rm nvidia/cuda:10.0-runtime-ubuntu18.04 nvidia-smi
Standard NVIDIA-SMI output shows the GPUs in your instance.
Installation
Download OmniSci from DockerHub and Start OmniSci in Docker.
sudo docker run --runtime=nvidia \ -d --runtime=nvidia \ -v /var/lib/omnisci:/omnisci-storage \ -p 6273-6280:6273-6280 \ omnisci/omnisci-ee-cuda:v5.3.0
See also the note regarding the CUDA JIT Cache in Optimizing Performance.
Ensure that you have sufficient storage on the drive you choose for /var/lib/omnisci.
| Note |
On startup, you receive the following error:
config file does not exist, ignoring --config /omnisci-storage/omnisci.confThis is expected behavior, because OmniSci does not ship with a default omnisci.conf file. To use OmniSci with the capabilities enabled by omnisci.conf:
|
For more information on CUDA driver installation, see the CUDA Installation Guide.
For more information on Docker installation, see the Docker Installation Guide.
Enter Your License Key
Validate your OmniSci instance with your license key.
- Copy your license key from the registration email message.
If you have not received your license key, contact your Sales Representative or register for your 30-day trial here. - Connect to Immerse using a web browser connected to your host machine on
port 6273. For example,
http://omnisci.mycompany.com:6273. - When prompted, paste your license key in the text box and click Apply.
- Click Connect to start using OmniSci.
Command Line Access
You can access the command line in the Docker image to perform configuration and run OmniSci utilities.
You need to know the container-id to access the command line. Use the command below to list the running containers.
docker container ls
You see output similar to the following.
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES 9e01e520c30c omnisci/omnisci-ee-cpu "/bin/sh -c '/omnisci..." 3 days ago Up 3 days 0.0.0.0:6273-6280->6273-6280/tcp confident_neumann
Once you have your container ID, you can access the command line using the
Docker exec command. For example, here is the command to start a Bash session
in the Docker instance listed above. The -it switch makes the
session interactive.
docker exec -it 9e01e520c30c bash
You can end the Bash session with the exit command.
Checkpoint
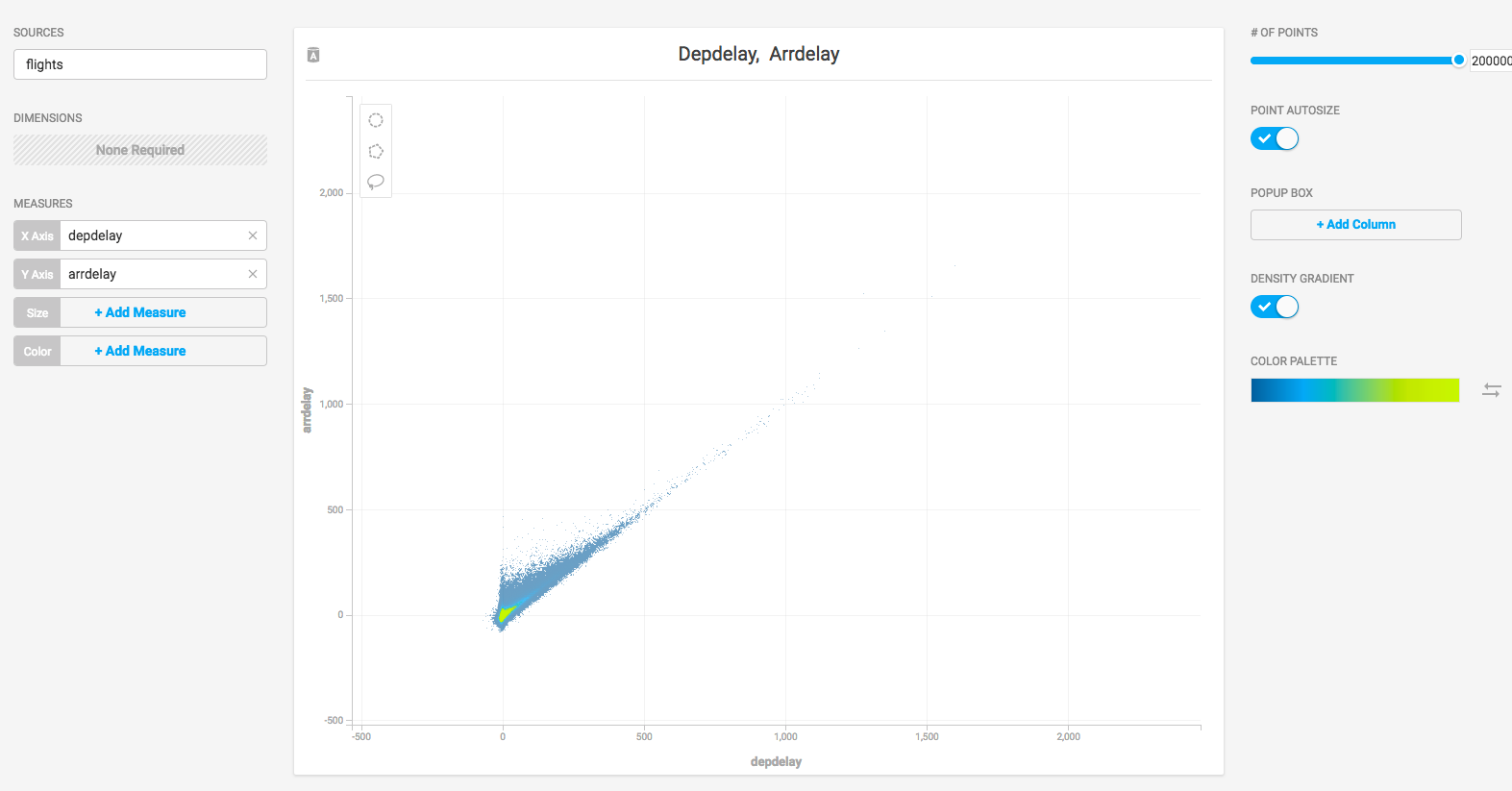
To verify that all systems are go, load some sample data, perform an omnisql query, and generate a pointmap using Immerse.
OmniSci ships with two sample datasets of airline flight information collected in 2008, and one dataset of New York City census information collected in 2015. To install the sample data, run the following command.
docker exec -it <container-id> ./insert_sample_data
Where <container-id> is the container in which OmniSci is running.
When prompted, choose whether to insert dataset 1 (7,000,000 rows), dataset 2 (10,000 rows), or dataset 3 (683,000 rows). The examples below use dataset 2.
Enter dataset number to download, or 'q' to quit:
# Dataset Rows Table Name File Name
1) Flights (2008) 7M flights_2008_7M flights_2008_7M.tar.gz
2) Flights (2008) 10k flights_2008_10k flights_2008_10k.tar.gz
3) NYC Tree Census (2015) 683k nyc_trees_2015_683k nyc_trees_2015_683k.tar.gzConnect to OmniSciDB by entering the following command (default password is HyperInteractive):
docker exec -it <container-id> /omnisci/bin/omnisql password: ••••••••••••••••
Enter a SQL query such as the following:
omnisql> SELECT origin_city AS "Origin", dest_city AS "Destination", AVG(airtime) AS "Average Airtime" FROM flights_2008_10k WHERE distance < 175 GROUP BY origin_city, dest_city;
The results should be similar to the results below.
Origin|Destination|Average Airtime Austin|Houston|33.055556 Norfolk|Baltimore|36.071429 Ft. Myers|Orlando|28.666667 Orlando|Ft. Myers|32.583333 Houston|Austin|29.611111 Baltimore|Norfolk|31.714286
Connect to Immerse using a web browser connected to your host machine on port 6273. For example, http://localhost:6273.
Create a new dashboard and a Scatter Plot to verify that backend rendering is working.
- Click New Dashboard.
- Click Add Chart.
- Click SCATTER.
- Click Select Data Source.
- Choose the flights_2008_10k table as the data source.
- Click X Axis +Add Measure.
- Choose depdelay.
- Click Y Axis +Add Measure.
- Choose arrdelay.
The resulting chart shows, unsurprisingly, that there is a correlation between departure delay and arrival delay.