Jupyter Integration
You can use JupyterLab to access OmniSciDB.
- Installing Jupyter with OmniSciDB
- Adding Jupyter to an Existing OmniSciDB Instance
- Creating the jhub_omnisci_dropbox Directory
- Upgrading docker-compose Services
- Using Jupyter
Installing Jupyter with OmniSciDB
| Note | When OmniSci is running in Docker, the paths used must be relative paths accessible to Docker, instead of OS-level paths. Keep the following in mind when performing Docker-based installations:
|
- Install the NVIDIA drivers and nvidia-container-runtime for your operating system, using the instructions at https://github.com/NVIDIA/nvidia-container-runtime.
- For Apt-based installations, such as Ubuntu, use the Docker preparation instructions.
- Change the default Docker runtime to
nvidiaand restart Docker.- Edit
/etc/docker/daemon.jsonand add"default-runtime": "nvidia". The resulting file should look similar to the following:{ "default-runtime": "nvidia", "runtimes": { "nvidia": { "path": "nvidia-container-runtime", "runtimeArgs": [] } } } - Restart the Docker service:
sudo systemctl restart docker
- Validate NVIDIA Docker:
docker run --rm nvidia/cudagl:10.1-runtime-ubuntu18.04 nvidia-smi
- Edit
- Install
docker-composeversion 1.25.1 or higher:sudo curl -L https://github.com/docker/compose/releases/download/1.25.1/docker-compose-`uname -s`-`uname -m` -o /usr/local/bin/docker-compose sudo chmod +x /usr/local/bin/docker-compose
- Create an OmniSci storage directory with a name of your choosing and change directories to it.
sudo mkdir /var/lib/omnisci cd /var/lib/omnisci
- Create the file
/var/lib/omnisci/omnisci.conf, and configure thejupyter-urlsetting under the[web]section to point to the Jupyter service:[web] jupyter-url = "http://jupyterhub:8000" servers-json = "/omnisci-storage/servers.json"
- Create the file
/var/lib/omnisci/data/omnisci.license. Copy your license key from the registration email message. If you have not received your license key, contact your Sales Representative or register for your 30-day trial here. - Create the following
/var/lib/omnisci/servers.jsonentry to enable Jupyter features in Immerse.[ { "enableJupyter": true } ] - Create
/var/lib/omnisci/docker-compose.yml:version: '3.7' services: omnisciserver: container_name: omnisciserver image: omnisci/omnisci-ee-cuda:v5.3.0 restart: always ipc: shareable volumes: - /var/lib/omnisci:/omnisci-storage - /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox:/jhub_omnisci_dropbox networks: - omnisci-frontend - omnisci-backend ports: - "6273:6273" - "6274:6274" - "6278:6278" # If using binary encryption, uncomment the below line to override the default # command that uses startomnisci, noting that you must have an existing OmniSci # data directory and run initdb before making this change. # command: /bin/bash -c "/omnisci/bin/omnisci_web_server --config /omnisci-storage/omnisci.conf & /omnisci/bin/omnisci_server --config /omnisci-storage/omnisci.conf" jupyterhub: container_name: jupyterhub image: omnisci/omnisci-jupyterhub:v0.3 restart: always networks: - omnisci-backend depends_on: - omnisciserver volumes: - /var/run/docker.sock:/var/run/docker.sock # Map this volume if binary encryption mode is configured and certificates are being validated # - /var/lib/omnisci/cacerts.crt:/omnisci-storage/cacerts.crt environment: ### Required settings ### # Note: if changing the image, do not remove the &jlabimage anchor # The gpu image includes Rapids, requires GPU devices and nvidia docker runtime: DOCKER_JUPYTER_LAB_IMAGE: &jlabimage omnisci/omnisci-jupyterlab-gpu:v0.3 # Without rapids does not require GPU: # DOCKER_JUPYTER_LAB_IMAGE: &jlabimage omnisci/omnisci-jupyterlab-cpu:v0.3 OMNISCI_HOST: omnisciserver OMNISCI_JUPYTER_ROLE: omnisci_jupyter ### Optional settings ### # DOCKER_NOTEBOOK_DIR: /home/jovyan # The directory inside the user's Jupyter Lab container to mount the user volume to. # HUB_IP: jupyterhub # The hostname or IP of the Jupyter Hub server # JHUB_BASE_URL: /jupyter/ # The base URL prepended to all Jupyter Hub and Lab requests # JHUB_ENABLE_ADMIN_ACCESS: "false" # Enable / disable admin access to Jupyter Hub # JLAB_CONTAINER_AUTOREMOVE: "true" # Enable / disable automatic removal of stopped Juptyer Lab containers # JLAB_DOCKER_NETWORK_NAME: omnisci-backend # The docker network name for Jupyter Lab containers # JLAB_IDLE_TIMEOUT: 3600 # Shut down Jupyter Lab containers after this many seconds of idle time # JLAB_NAME_PREFIX: jupyterlab # Container name prefix for Lab containers # JLAB_OMNISCI_IMPORT_VOLUME_PATH: /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox # Local Docker host path for where to mount the shared directory available to the OmniSciDB server for file ingest # JLAB_NOTEBOOK_TERMINALS_ENBLED: "false" # Enable terminals in notebooks # JLAB_USER_VOLUME_PATH: /var/lib/omnisci/jupyter/ # Local Docker host path to be used for user Jupyter Lab home directory volume mapping # JUPYTER_DEBUG: "true" # Turn on / off debugging for Jupyter Hub and Lab # OMNISCI_ALLOW_SUPERUSER_ROLE: "false" # Authorize super users access to Jupyter without needing to be given the OmniSci role specified in OMNISCI_JUPYTER_ROLE # OMNISCI_BINARY_TLS_CACERTS_LOCAL_PATH: /var/lib/omnisci/cacerts.crt # Specifying this or mapping a volume in the Hub container to /omnisci-storage/cacerts.crt will automatically enable binary TLS mode # OMNISCI_BINARY_TLS_VALIDATE: "false" # Whether or not to validate certificates in binary TLS mode. Specifying either "true" or "false" will enable binary TLS mode # OMNISCI_PORT: 6278 # Port that Jupyter Hub should use to connect to OmniSci. Ensure this matches the protocol # OMNISCI_PROTOCOL: http # Protocol that Jupyter Hub should use to connect to OmniSci. Ensure this is "binary" if using binary encryption # SPAWNER_CPU_LIMIT: 1 # Number of CPU cores available for each Jupyter Lab container, see https://jupyterhub.readthedocs.io/en/stable/api/spawner.html#jupyterhub.spawner.Spawner.cpu_limit # SPAWNER_RAM_LIMIT: 10G # Amount of CPU RAM available for each Jupyter Lab container, see https://jupyterhub.readthedocs.io/en/stable/api/spawner.html#jupyterhub.spawner.LocalProcessSpawner.mem_limit # SPAWNER_ENV_OMNISCI_HOST: omnisciserver # Hostname / IP address of the OmniSci server for Lab containers to connect to by default # SPAWNER_ENV_OMNISCI_PORT: 6274 # Port of the OmniSci server for Lab containers to connect to by default # SPAWNER_ENV_OMNISCI_PROTOCOL: binary # Protocol of the OmniSci server for Lab containers to connect to by default # OMNISCI_DB_URL: "omnisci://omnisciserver:6274/omnisci" # Alternative, direct connection (not Immerse session) to omnisciserver. Username will be inferred, but password will be required in notebook. # More volumes for lab containers: # JLAB_VOLUME_1: /data1:/data1:rw # JLAB_VOLUME_2: /var/lib/omnisci:/omnisci-storage:ro # The purpose of this is to make sure the jlabimage is pulled because jupyterhub will not pull it automatically when launching jupyterlab-tmp: image: *jlabimage command: echo networks: - omnisci-backend networks: omnisci-frontend: driver: bridge name: omnisci-frontend omnisci-backend: driver: bridge name: omnisci-backend - Run
docker-compose pullto download the images from Docker Hub. Optional when you specify a version indocker-compose.yml. - Make sure you are in the directory that you created in step 5, and run
composein detached mode:docker-compose up -d
composerestarts by default whenever stopped. - Log in as the super user (
admin/HyperInteractive). - Create required users in OmniSciDB.
- Create the omnisci_jupyter role in OmniSciDB:
CREATE ROLE omnisci_jupyter
- Grant the omnisci_jupyter role to users who require Jupyter access:
GRANT omnisci_jupyter TO username
| Note | You might need to stop lab containers for them to be restarted with the new image:
docker ps | grep jupyterlab docker rm -f jupyterlab-USERNAME |
You should now see Jupyter icons in the upper right of Immerse and when running queries in SQL Editor.
Adding Jupyter to an Existing OmniSciDB Instance
| Note | When OmniSci is running in Docker, the paths used must be relative paths accessible to Docker, intead of OS-level paths. Keep the following in mind when peforming Docker-based installations:
|
To use Jupyter with an existing, non-Docker install of OmniSci, change OmniSci to run on Docker instead of the host. The steps are the same as the install instructions, with the following exceptions:
- Change the volume mappings highlighted in bold to point to your existing installation path:
omnisciserver: container_name: omnisciserver image: omnisci/omnisci-ee-cuda:v5.3.0 restart: always ipc: shareable volumes: - /var/lib/omnisci:/omnisci-storage - /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox:/jhub_omnisci_dropbox - Enable the following environment variables and change the relevant paths,
highlighted in bold, to your existing installation:
### Optional settings ### # DOCKER_NOTEBOOK_DIR: /home/jovyan # The directory inside the user's Jupyter Lab container to mount the user volume to. # HUB_IP: jupyterhub # The hostname or IP of the Jupyter Hub server # JHUB_BASE_URL: /jupyter/ # The base URL prepended to all Jupyter Hub and Lab requests # JHUB_ENABLE_ADMIN_ACCESS: "false" # Enable / disable admin access to Jupyter Hub # JLAB_DOCKER_NETWORK_NAME: omnisci-backend # The docker network name for Jupyter Lab containers # JLAB_IDLE_TIMEOUT: 3600 # Shut down Jupyter Lab containers after this many seconds of idle time # JLAB_NAME_PREFIX: jupyterlab # Container name prefix for Lab containers JLAB_OMNISCI_IMPORT_VOLUME_PATH: /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox # Local Docker host path for where to mount the shared directory available to the OmniSciDB server for file ingest # JLAB_NOTEBOOK_TERMINALS_ENBLED: "false" # Enable terminals in notebooks JLAB_USER_VOLUME_PATH: /var/lib/omnisci/jupyter/ # Local Docker host path to be used for user Jupyter Lab home directory volume mapping # JUPYTER_DEBUG: "true" # Turn on / off debugging for Jupyter Hub and Lab # OMNISCIDB_CONTAINER_NAME: omnisciserver # OmniSciDB container name for IPC sharing with Lab containers # SPAWNER_CPU_LIMIT: 1 # Number of CPU cores available for each Jupyter Lab container, see https://jupyterhub.readthedocs.io/en/stable/api/spawner.html#jupyterhub.spawner.Spawner.cpu_limit # SPAWNER_RAM_LIMIT: 10G # Amount of CPU RAM available for each Jupyter Lab container, see https://jupyterhub.readthedocs.io/en/stable/api/spawner.html#jupyterhub.spawner.LocalProcessSpawner.mem_limit # SPAWNER_ENV_OMNISCI_HOST: omnisciserver # Hostname / IP address of the OmniSci server for Lab containers to connect to by default # SPAWNER_ENV_OMNISCI_PORT: 6274 # Port of the OmniSci server for Lab containers to connect to by default # SPAWNER_ENV_OMNISCI_PROTOCOL: binary # Protocol of the OmniSci server for Lab containers to connect to by default - If you have an existing omnisci.conf file:
- Add the required sections
instead of creating a new file:
[web] jupyter-url = "http://jupyterhub:8000" servers-json = "/omnisci-storage/servers.json"
- Remove the data, port, http-port, and frontend properties, which should not be changed with a Docker installation.
- Ensure that all paths, such as cert and key, are accessible by Docker.
- Add the required sections
instead of creating a new file:
- If you have an existing servers.json file, move it to your OmniSci
home directory (/var/lib/omnisci by default) and add the following key/value
pair:
"enableJupyter": true
| Important | Before running docker-compose up -d, ensure that any existing
installations are stopped and disabled. For example:
sudo systemctl stop omnisci_server sudo systemctl stop omnisci_web_server sudo systemctl disable omnisci_server sudo systemctl disable omnisci_web_serverAs with any software upgrade, back up your data before you upgrade OmniSci. |
Creating the jhub_omnisci_dropbox Directory
Run the following commands to create the jhub_omnisci_dropbox directory and make it writeable by your users. Change the volume mappings highlighted in bold to point to your existing installation path:
sudo mkdir /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox sudo chown 1000 /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox sudo chmod 750 /var/lib/omnisci/data/omnisci_import/jhub_omnisci_dropbox
This allows Jupyter users to write files into /home/jovyan/jhub_omnisci_dropbox/.
You can also use that directory path in COPY FROM SQL commands.
Upgrading docker-compose Services
To upgrade Jupyter images using the docker-compose.yml file, edit docker-compose.yml as follows:
services:
omnisciserver:
...
image: omnisci/omnisci-ee-cuda:v5.3.0
...
jupyterhub:
container_name: jupyterhub
image: omnisci/omnisci-jupyterhub:v0.3
...
environment:
...
DOCKER_JUPYTER_LAB_IMAGE: &jlabimage omnisci/omnisci-jupyterlab-gpu:v0.3
Then, use the following commands to download the images and restart the services with the new versions:
docker-compose pull docker-compose up -d
You might also need to stop lab containers in order to start them again in a new image:
docker ps | grep jupyterlab
For each user, run the following command:
docker rm -f jupyterlab-<username>
Using Jupyter
Open JupyterLab by clicking the Jupyter icon in the upper right corner of Immerse.
JupyterLab opens in a new tab. You are signed in automatically using OmniSci authentication, with a notebook ready to start an OmniSci connection. The notebook is saved for you at the root of your Jupyter file system.
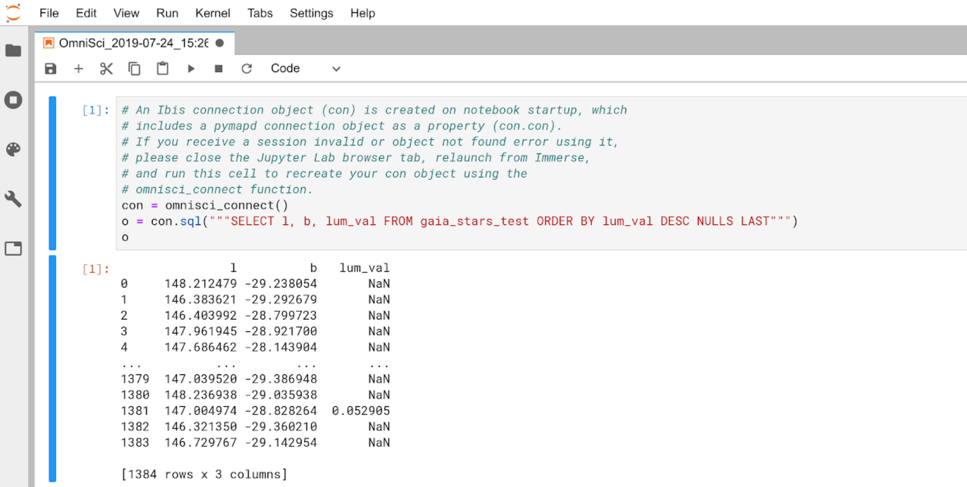
You can verify the location of your file by clicking the folder icon in the top left.
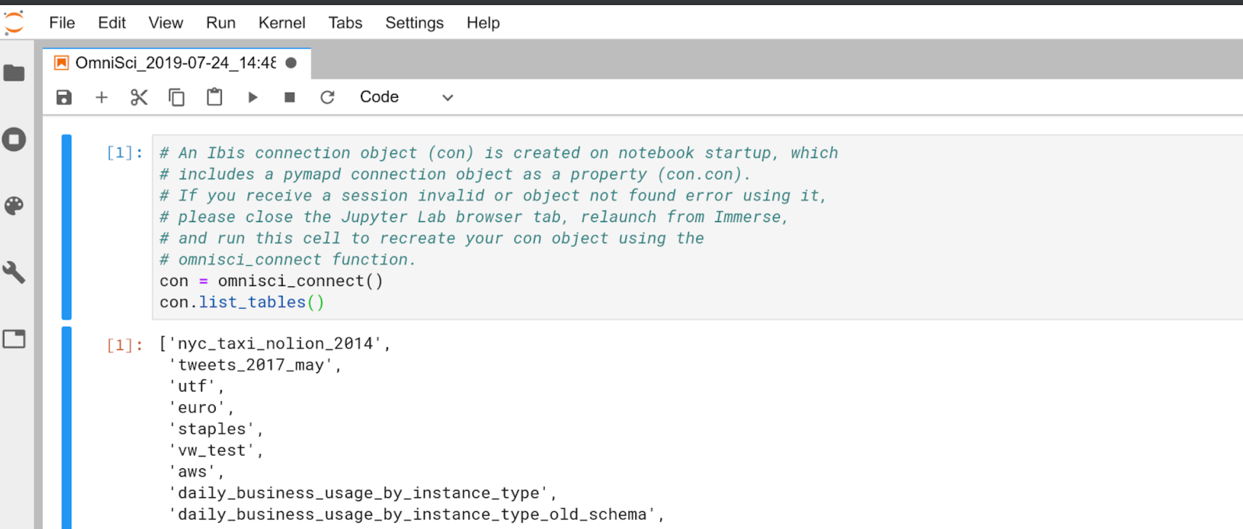
The contents of the cell are prefilled with explanatory comments and the
omnisci_connect() method ready to set up your Omnisci connection
in Jupyter. Click the Play button to run the connection statement
and list the tables in your OmniSciDB instance.
You can continue to use con to run more Ibis expressions in other
cells.
The connection reuses the session already in use by Immerse by passing Jupyter the raw session ID. If you connect this way without credentials, the connection has a time-to-live (TTL). After the session timeout period passes with no activity (60 minutes by default), the session is invalidated. You have to reenter Jupyter from Immerse in the same way to reestablish the connection, or use the pymapd connect method to enter your credentials manually.
You can also launch Jupyter from the Immerse SQL Editor. After you run a query in the SQL Editor, you see a button that allows you to send your query to Jupyter.
The query displays in a different notebook, ready to run the query. You must run the cell yourself to send the query and see the results.